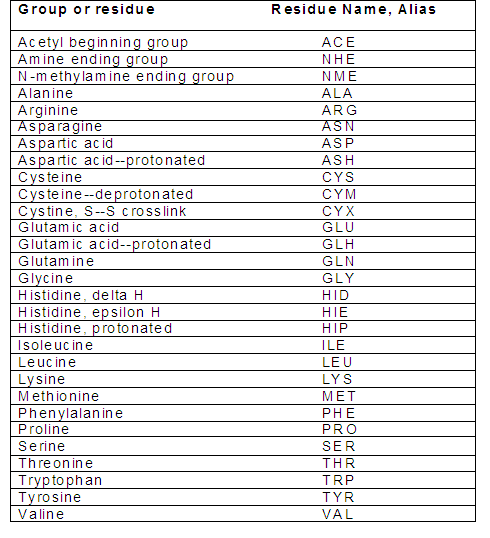
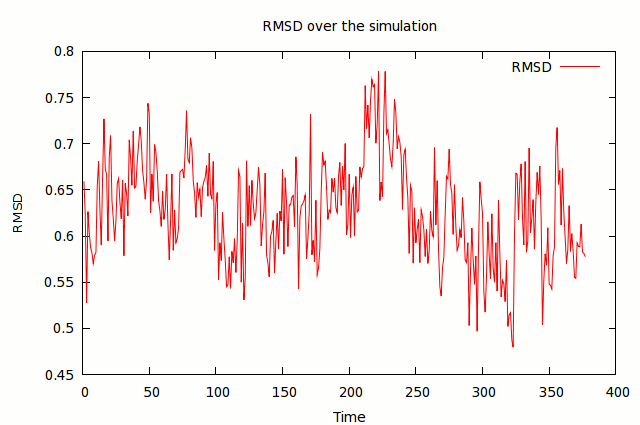
amber 19 computational chemistry chemistry

open babel an open chemical toolbox journal of cheminformatics full text

charmm gui pdb manipulator for advanced modeling and simulations of proteins containing nonstandard residues sciencedirect

zaygs7966aqdnm

chemical formats labii eln lims

290 questions with answers in pymol science topic

mercury crystallography wikipedia

studylib

getting started isolde 1 0b3 documentation

polypargen

tripos mol2 file format amber mailing list archive

ucsf dock

tripos mol2 file format amber mailing list archive

ucsf dock

cpptraj manual

molecule viewer macs in chemistry

cpptraj manual

ambertools users manual

pdf amber 9 university of california san francisco

glide docking tutorial docking on an rna receptor amber chpc wiki

automated generation of mcss derived pharmacophoric dock site points for searching multiconformation databases joseph mccarthy 2003 proteins structure function and bioinformatics wiley online library

automated generation of mcss derived pharmacophoric dock site points for searching multiconformation databases joseph mccarthy 2003 proteins structure function and bioinformatics wiley online library

researchgate

preparing your system for molecular dynamics md biochemcore 2018

frontiers e bitter bitterant prediction by the consensus voting from the machine learning methods chemistry
You May Like